NcoI
2025-01-23
Recombinant Ribonuclease A (RNase A) Solution (Animal free)
2025-01-23HAEIII
Product Number: HAE01
Shipping and Storage
Stored at -20ºC, valid for two years.
Components
| Component | HAE01 400μl |
| HAEIII | 400μl |
| 10×Cut Buffer | 1ml |
| Easy-Load 10×Cut Buffer | 1ml |
Description
HAEIII is a high-quality restriction endonuclease that has been genetically engineered and can quickly complete DNA cleavage using only one buffer within 5-15 minutes.Suitable for rapid enzymatic digestion of plasmid DNA, PCR products, or genomic DNA.
- Enzyme activity detection: At the optimal reaction temperature, in a 20μl reaction system, 1μl of HaeIII can completely digest 1μg of λ DNA containing 149 HaeIII enzyme cleavage sites within 15 minutes.
- Long term enzyme digestion detection: Incubate 1μl HAEIII with 1μg λDNA for 3 hours at the optimal reaction temperature, and no non-specific degradation of the substrate caused by other nucleases contamination or star activity was detected. Delayed enzyme digestion may result in star activity.
- Enzyme digestion ligation re digestion detection: At the optimal reaction temperature, use 1μl HAEIII to digest the substrate, recover the enzyme digestion product, and use an appropriate amount of T4 DNA Ligase at 22℃ to reconnect the enzyme digestion product. After recovering the ligation product again, use the same endonuclease to cleave the ligation product again.
- Detection of non-specific endonuclease activity: at the optimal reaction temperature, 1μl HAEIII and 1μg super spiral plasmid DNA were incubated together for 4h, and agarose gel electrophoresis was used to detect that the plasmid DNA was still in the super spiral state.
- Blue and white spot detection: The vector containing a single lacZ α gene was digested with 1μl HAEIII, reconnected, and transformed into competent E. coli cells, and coated on LB medium plates containing corresponding antibiotics, IPTG, and X-gal. Products with correct connections will grow blue colonies, while products with incorrect connections (i.e. incomplete DNA end incisions) will grow white colonies. For HAEIII, the proportion of white colonies should be less than 1%.
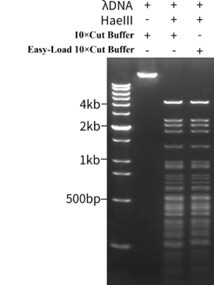
Figure 1. Experimental results of HaeIII enzyme activity. A 20μl reaction system containing 1μg of λ DNA (48.5kb) and 1μl of HaeIII without or with the addition of 1X Cut Buffer and 1X Easy Load Cut Buffer, respectively, was incubated at 37°C for 15 minutes for enzyme cleavage reaction, followed by incubation at 80°C for 20 minutes to inactivate the enzyme, followed by electrophoresis and Gel Red (EB upgraded product), Perform nucleic acid staining and fluorescence imaging analysis at 10000X. The DNA marker used is DNA Ladder (0.2-12 kb, 12 bands). The actual detection effect may vary due to differences in experimental conditions, detection instruments, etc. The effect shown in the figure is for reference only.
Basic information
| Recognition sequence | Isoschizomer | Enzyme digestion temperature | Deactivation conditions | Methylation interference? |
| 5′-GG^CC-3′ 3′-CC^GG-5′ | BshFI, BsnI, BspANI, BsuRI | 37℃ | 80ºC 20min | None |
The activity (buffer compatibility) in different reaction buffers is as follows:
| 10×Cut Buffer | Easy-Load 10×Cut Buffer | Thermo FastDigest Buffer | NEB CutSmart® Buffer | Takara QuickCut™ Buffer |
| 100% | 100% | 100% | 100% | 100% |
Please refer to the table below for the methylation effects of HAEIII recognition sites:
| Dam | Dcm | CpG | EcoKI | EcoBI |
| No effect | No effect | No effect | No effect | No effect |
Features
- Enzymatic cleavage can be completed within 5-15 minutes;
- All endonucleases share a single enzyme digestion buffer, Cut Buffer, greatly simplifying the enzyme digestion reaction system and facilitating double or multiple enzyme digestion;
- In response to the issue of differences in activity of different enzymes in Cut Buffer, the concentrations of different enzymes were adjusted to uniformly add 1μl of enzyme per 20μl of system for enzyme digestion reaction;
- Many modifying enzymes, such as Alkaline Phosphatase, Antarctic Phosphatase, T4 DNA Ligase, T4 Polynucleotide Kinase, T4 PNK (3 ‘phosphatase minus), etc., are 100% compatible with Cut Buffer, making reaction systems such as “enzyme cut connect” and “enzyme cut modify connect” compatible and supporting single tube reactions;
- Good enzyme activity redundancy makes it easy to cope with substrate excess or difficult template enzyme digestion.